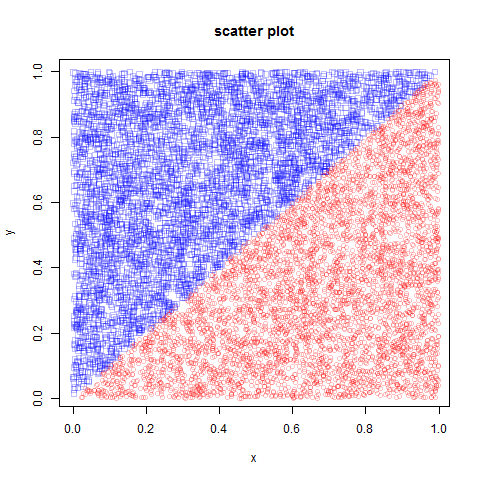
create training data:
n=10000 x=runif(n) y=runif(n) df=data.frame(x,y,as.factor(x>y))
visualize:
colors = list(rgb(0,0,1,1/4),rgb(1,0,0,1/4))
names(df) = c("x", "y", "class")
classes=levels(df[[3]])
obs1=as.matrix(subset(df,class==classes[[1]])[,1:2])
obs2=as.matrix(subset(df,class==classes[[2]])[,1:2])
# make scatter plot
dev.new()
plot(obs1[,1],obs1[,2],xlab="x",ylab="y",main="scatter plot",pch=0,col=colors[[1]])
points(obs2[,1],obs2[,2],xlab="x",ylab="y",main="scatter plot",pch=1,col=colors[[2]])
result:
D = \frac{|\mu_1-\mu_2|}{\sigma_1+\sigma_2}:
> Dp
x y
0.7121875 0.7208857
build classifier:
model=glm(class~.,family="binomial",data=df) summary(model) # prints summary
Output:
Call:
glm(formula = class ~ ., family = "binomial", data = df)
Deviance Residuals:
Min 1Q Median 3Q Max
-0.11832 0.00000 0.00000 0.00000 0.08847
Coefficients:
Estimate Std. Error z value Pr(>|z|)
(Intercept) 5.765e-01 1.923e+01 0.030 0.976
feature1 9.761e+04 8.981e+04 1.087 0.277
feature2 -9.761e+04 8.981e+04 -1.087 0.277
(Dispersion parameter for binomial family taken to be 1)
Null deviance: 1.3863e+04 on 9999 degrees of freedom
Residual deviance: 2.9418e-02 on 9997 degrees of freedom
AIC: 6.0294
Number of Fisher Scoring iterations: 25
algorithm fails to converge because of perfect separation:
Warning messages: 1: glm.fit: algorithm did not converge 2: glm.fit: fitted probabilities numerically 0 or 1 occurred
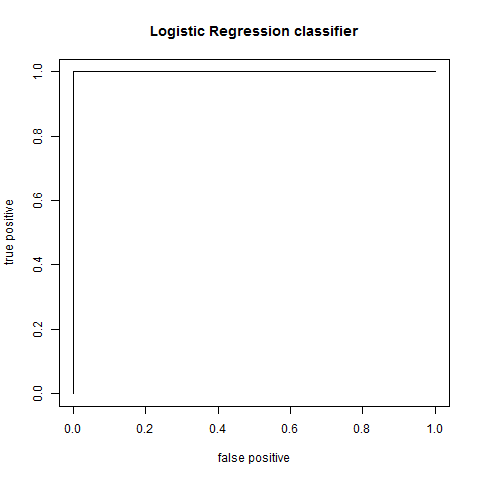
compute AUC:
response=predict(model,type="response")
groundTruth=df[,3]
M=NULL
for (threshold in seq(0.05,0.95,0.05))
{
M=cbind(M,calc(response,groundTruth,classes,threshold))
}
M = cbind(c(1,1),M,c(0,0))
fp = M[1,]
tp = M[2,]
auc = -auc(fp, tp)
where:
auc=function(x,y)
{
d = diff(x, 1)
e = as.numeric(lapply(1:(length(y)-1), function(i)y[i]+y[i+1]))
return(sum(0.5*d*e))
}
calc=function(response, groundTruth, classes, threshold)
{
type1 = classes[1]
type2 = classes[2]
n=length(response)
predicted = as.factor(ifelse(response<threshold,type1,type2))
I = which(groundTruth==type1)
fp = length(which(predicted[I] != type1)) / length(I)
I = which(groundTruth==type2)
tp = length(which(predicted[I] == type2)) / length(I)
return(c(fp,tp))
}
plot AUC:
plot(fp,tp,type="l",xlab="false positive",ylab="true positive",main="Logistic Regression classifier")
result:
LDA:
lda_classifier=lda(class~., data=df)
Call:
lda(class ~ ., data = df)
Prior probabilities of groups:
FALSE TRUE
0.5007 0.4993
Group means:
feature1 feature2
FALSE 0.3346288 0.6676169
TRUE 0.6710111 0.3380432
Coefficients of linear discriminants:
LD1
**feature1 4.280490
feature2 -4.196388**
finally you add add some noise to make logistic regression converge:
df=data.frame(x+rnorm(n,sd=0.1),y+rnorm(n,sd=0.1),as.factor(x>y))




